PUBLICATIONS
Relevant articles
Johann Mertens, Santiago Casado, Carlos P. Mata, Mercedes Hernando-Pérez, Pedro J. de Pablo, José L. Carrascosa & José R. Castón
Viral capsids are metastable structures that perform many essential processes; they also act as robust cages during the extracellular phase. Viruses can use multifunctional proteins to optimize resources (e.g., VP3 in avian infectious bursal disease virus, IBDV). The IBDV genome is organized as ribonucleoproteins (RNP) of dsRNA with VP3, which also acts as a scaffold during capsid assembly. We characterized mechanical properties of IBDV populations with different RNP content (ranging from none to four RNP). The IBDV population with the greatest RNP number (and best fitness) showed greatest capsid rigidity. When bound to dsRNA, VP3 reinforces virus stiffness. These contacts involve interactions with capsid structural subunits that differ from the initial interactions during capsid assembly. Our results suggest that RNP dimers are the basic stabilization units of the virion, provide better understanding of multifunctional proteins, and highlight the duality of RNP as capsid-stabilizing and genetic information platforms.
Verónica A. González-García, Mar Pulido-Cid, Carmela Garcia-Doval, Rebeca Bocanegra, Mark J. van Raaij, Jaime Martín-Benito, Ana Cuervo and José L. Carrascosa
The majority of bacteriophages protect their genetic material by packaging the nucleic acid in concentric layers to an almost crystalline concentration inside protein shells (capsid). This highly condensed genome also has to be efficiently injected into the host bacterium in a process named ejection. Most phages use a specialized complex (often a tail) to deliver the genome without disrupting cell integrity. Bacteriophage T7 belongs to the Podoviridae family and has a short, non-contractile tail formed by a tubular structure surrounded by fibers. Here we characterize the kinetics and structure of bacteriophage T7 DNA delivery process. We show that T7 recognizes lipopolysaccharides (LPS) from Escherichia coli rough strains through the fibers. Rough LPS acts as the main phage receptor and drives DNA ejection in vitro. The structural characterization of the phage tail after ejection using cryo-electron microscopy (cryo-EM) and single particle reconstruction methods revealed the major conformational changes needed for DNA delivery at low resolution. Interaction with the receptor causes fiber tilting and opening of the internal tail channel by untwisting the nozzle domain, allowing release of DNA and probably of the internal head proteins.
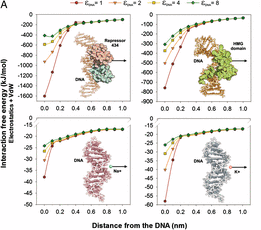
Ana Cuervo, Pablo D. Dans, José L. Carrascosa, Modesto Orozco, Gabriel Gomila and Laura Fumagalli
The electric polarizability of DNA, represented by the dielectric constant, is a key intrinsic property that modulates DNA interaction with effector proteins. Surprisingly, it has so far remained unknown owing to the lack of experimental tools able to access it. Here, we experimentally resolved it by detecting the ultraweak polarization forces of DNA inside single T7 bacteriophages particles using electrostatic force microscopy. In contrast to the common assumption of low-polarizable behavior like proteins (εr ∼ 2–4), we found that the DNA dielectric constant is ∼8, considerably higher than the value of ∼3 found for capsid proteins. State-of-the-art molecular dynamic simulations confirm the experimental findings, which result in sensibly decreased DNA interaction free energy than normally predicted by Poisson–Boltzmann methods. Our findings reveal a property at the basis of DNA structure and functions that is needed for realistic theoretical descriptions, and illustrate the synergetic power of scanning probe microscopy and theoretical computation techniques.
Mercedes Hernando-Pérez, Elena Pascual, María Aznar, Alina Ionel José R. Castón, Antoni Luque, José L. Carrascosa, David Reguera and Pedro J. de Pablo
The stability and strength of viral nanoparticles are crucial to fulfill the functions required through the viral cycle as well as using capsids for biomedical and nanotechnological applications. The mechanical properties of viral shells obtained through Atomic Force Microscopy (AFM) and continuum elasticity theory, such as stiffness or Young's modulus, have been interpreted very often in terms of stability. However, viruses are normally subjected to chemical rather than to mechanical aggression. Thus, a correct interpretation of mechanics in terms of stability requires an adequate linkage between the ability of viral cages to support chemical and mechanical stresses. Here we study the mechanical fragility and chemical stability of bacteriophage T7 in two different maturation states: the early proheads and the final mature capsids. Using chemical stress experiments we show that proheads are less stable than final mature capsids. Still, both particles present similar anisotropic stiffness, indicating that a continuum elasticity description in terms of Young's modulus is not an adequate measure of viral stability. In combination with a computational coarse-grained model we demonstrate that mechanical anisotropy of T7 emerges out of the discrete nature of the proheads and empty capsids. Even though they present the same stiffness, proheads break earlier and have fractures ten times larger than mature capsids, in agreement with chemical stability, thus demonstrating that fragility rather than stiffness is a better indicator of viral cages' stability.
Last 5 years
237
Joaquín Otón, Eva Pereiro, José J. Conesa, Francisco J. Chichón, Daniel Luque, Javier M. Rodríguez, Ana J. Pérez-Berná, Carlos Oscar S. Sorzano, Joanna Klukowska, Gabor T. Herman, Javier Vargas, Roberto Marabini, José L. Carrascosa & José M. Carazo
XTEND: Extending the depth of field in cryo soft X-ray tomography.
Scientific Reports, 7, Article number: 45808, doi: 10.1038/srep45808 (2017)
236
Maria Rosario Fernandez-Fernandez, Desire Ruiz-Garcia, Eva Martin-Solana, Francisco Javier Chichon, Jose L. Carrascosa, Jose-Jesus Fernandez.
3D electron tomography of brain tissue unveils distinct Golgi structures that sequester cytoplasmic contents in neurons.
J Cell Sci, 130, 83-89, doi: 10.1242/jcs.188060 (2017)
235
Ramírez-Santiago G, Robles-Valero J, Morlino G, Cruz-Adalia A, Pérez-Martínez M, Zaldivar A, Torres-Torresano M, Chichón FJ, Sorrentino A, Pereiro E, Carrascosa JL, Megías D, Sorzano CO, Sánchez-Madrid F, Veiga E.
Clathrin regulates lymphocyte migration by driving actin accumulation at the cellular leading edge.
Eur J Immunol, 46, 2376-2387, doi: 10.1002/eji.201646291 (2016)
234
Ana Joaquina Pérez-Berná, Maria José Rodríguez, Francisco Javier Chichón, Martina Friederike Friesland, Andrea Sorrentino, Jose L. Carrascosa, Eva Pereiro and Pablo Gastaminza.
Structural Changes In Cells Imaged by Soft X-ray Cryo-Tomography During Hepatitis C Virus Infection.
ACS Nano, doi: 10.1021/acsnano.6b01374 (2016)
233
Michele Chiappi, José Javier Conesa, Eva Pereiro, Carlos Oscar Sánchez Sorzano, María Josefa Rodríguez, Katja Henzler, Gerd Schneider, Francisco Javier Chichón and José L. Carrascosa.
Cryo-soft X-ray tomography as a quantitative three-dimensional tool to model nanoparticle:cell interaction.
Journal of Nanobiotechnology, 14:15 doi: 10.1186/s12951-016-0170-4 (2016)
232
Antonio Aires, Sandra M Ocampo, Bruno M Simões, María Josefa Rodríguez, Jael F Cadenas, Pierre Couleaud,
Katherine Spence, Alfonso Latorre, Rodolfo Miranda, Álvaro Somoza, Robert B Clarke, José L Carrascosa and
Aitziber L Cortajarena
Multifunctionalized iron oxide nanoparticles for selective drug delivery to CD44-positive cancer cells.
Nanotechnology, 27:6, 065103 doi: 10.1088/0957-4484/27/6/065103 (2016)
231
Jose Javier Conesa, Joaquín Otón, Michele Chiappi, Jose María Carazo, Eva Pereiro, Francisco Javier Chichón and José L. Carrascosa.
Intracellular nanoparticles mass quantification by near-edge absorption soft X-ray nanotomography.
Scientific Reports, 6:22354 doi: 10.1038/srep22354 (2016)
230
Miguel Ángel Cuesta-Geijo, Michele Chiappi, Inmaculada Galindo, Lucía Barrado-Gil, Raquel Muñoz-Moreno, José L. Carrascosa and Covadonga Alonso
Cholesterol flux is required for endosomal progression of African swine fever virions during the initial establishment of infection.
Journal of Virology, 90:3, 1534-1543, doi:10.1128/JVI.02694-15 (2016)
229
Sandra M. Ocampo, Vanessa Rodriguez, Leonor de la Cueva, Gorka Salas, José L. Carrascosa, María Josefa Rodríguez, Noemí García-Romero, Jose Luis F. Cuñado, Julio Camarero, Rodolfo Miranda, Cristobal Belda-Iniesta and Angel Ayuso-Sacido.
g-force induced giant efficiency of nanoparticles internalization into living cells.
Scientific Reports, 5:15160, doi: 10.1038/srep15160 (2015)
Impact Factor: 5.228 - Q1
228
Aneesh Vijayan, Juan García-Arriaza, Suresh C. Raman, José Javier Conesa, Francisco Javier Chichón, César Santiago, Carlos Óscar S. Sorzano, José L. Carrascosa and Mariano Esteban.
A Chimeric HIV-1 gp120 Fused with Vaccinia Virus 14K (A27) Protein as an HIV Immunogen.
PLoS One, 10:7, e0133595, doi: 10.1371/journal.pone.0133595 (2015)
Impact Factor: 3.057 - Q1
227
José M. Valpuesta and José L. Carrascosa
Electron microscopy: the coming of age of a structural biology technique.
Archives of Biochemistry and Biophysics, 581,1-2 doi: 10.1016/j.abb.2015.06.018 (2015)
Impact Factor: 2.807 – Q2
226
Johann Mertens, Santiago Casado, Carlos P. Mata, Mercedes Hernando-Pérez, Pedro J. de Pablo, José L. Carrascosa and José R. Castón
A protein with simultaneous capsid scaffolding and dsRNA-binding activities enhances the birnavirus capsid mechanical stability.
Scientific Reports, 5:13486, doi: 10.1038/srep13486 (2015)
Impact Factor: 5.228 - Q1
225
Verónica A. González-García, Rebeca Bocanegra, Mar Pulido-Cid, Jaime Martín-Benito, Ana Cuervo and José L. Carrascosa
Characterization of the initial steps in the T7 DNA ejection process.
Bacteriophage, vol. 5:3, doi: 10.1080/21597081.2015.1056904 (2015)
224
Ana J. Pérez-Berná, Sanjin Marion, F.Javier Chichón, José J. Fernández, Dennis C. Winkler, José L. Carrascosa, Alasdair C. Steven, Antonio Siber and Carmen San Martín
Distribution of DNA-condensing protein complexes in the adenovirus core.
Nucleic Acids Research, 43:8, 4274-4283, doi: 10.1093/nar/gkv187 (2015)
Impact Factor: 9.202 - Q1 – D1
223
Macarena Calero, Michele Chiappi, Ana Lazaro-Carrillo, María José Rodríguez, Francisco Javier Chichón, Kieran Crosbie-Staunton, Adriele Prina-Mello, Yuri Volkov, Angeles Villanueva and José L. Carrascosa
Characterization of interaction of magnetic nanoparticles with breast cancer cells.
Journal of Nanobiotechnology, 13:16, doi: 10.1186/s12951-015-0073-9 (2015)
Impact Factor: 4.239 - Q1
222
Verónica A. González-García, Mar Pulido Cid, Carmela García-Doval, Rebeca Bocanegra, Mark J. van Raaij, Jaime Martín-Benito, Ana Cuervo and José L. Carrascosa
Conformational Changes Leading to T7 DNA Delivery upon Interaction with the Bacterial Receptor.PORTADA
Journal of Biological Chemistry, 290:16, 10038-10044, doi: 10.1074/jbc.M114.614222 (2015)
Impact Factor: 4.258 - Q1
221
José A. Morín, Francisco J. Cao, José M. Lazaro, J. Ricardo Arias-González, José M. Valpuesta, José L. Carrascosa, Margarita Salas and Borja Ibarra
Mechano-chemical kinetics of DNA replication: identification of the translocation step of a replicative DNA polymerase.
Nucleic Acids Research, 43:7, 3643-3652, doi: 10.1093/nar/gkv204 (2015)
Impact Factor: 9.202 - Q1 – D1
220
Javier Lopez-Andarias, María José Rodríguez, Carmen Atienza, Juan Luis López, Tsubasa Mikie, Santiago Casado, Shu Seki, José L. Carrascosa and Nazario Martín.
Highly Ordered n/p-Co-assembled Materials with Remarkable Charge Mobilities
Journal of American Chemical Society, 137:2, 893−897, doi: 10.1021/ja510946c (2015)
Impact Factor: 13.038 - Q1 – D1
219
Rebecca L. Plimpton, Jorge Cuéllar, Chun Wan J. Lai, Takuma Aoba, Aman Makaju, Sarah Franklin, Andrew D. Mathis, John T. Prince, José L. Carrascosa, José M. Valpuesta, Barry M. Willardson.
Structures of the G‑CCT and PhLP1‑G‑CCT complexes reveal a mechanism for G-protein -subunit folding and Gγ dimer assembly.
Proc Natl Acad Sci USA, 112:8, 2413-2418, doi: 10.1073/pnas.1419595112 (2015)
Impact Factor: 9.423 - Q1 – D1
218
Elena Pascual, Carlos P. Mata, Josué Gómez-Blanco, Noelia Moreno, Juan Bárcena, Esther Blanco, Ariel Rodríguez-Frandsen, Amelia Nieto, José L. Carrascosa, José R.Castón
Structural Basis for the Development of Avian Virus Capsids That Display Influenza Virus Proteins and Induce Protective Immunity.
Journal of Virology, 89:5, 2563-2574 doi:10.1128/JVI.03025-14 (2015)
Impact Factor: 4.606 - Q1
217
Ana Cuervo, Pablo D. Dans, José L. Carrascosa, Modesto Orozco, Gabriel Gomila and Laura Fumagalli.
Direct measurement of the dielectric polarization properties or DNA.
Proc Natl Acad Sci USA, 111:35, E3624-E3630, doi: 10.1073/pnas.1405702111 (2014)
Impact Factor: 9.674 - Q1 – D1
216
Javier M. Rodríguez, Francisco J. Chichón, Esther Martín-Forero, Fernando González-Camacho, José L. Carrascosa, José R. Castón, Daniel Luque.
New Insights into Rotavirus Entry Machinery: Stabilization of Rotavirus Spike Conformation Is Independent of Trypsin Cleavage.
PLOS Pathogens, 10:5, e1004157, doi:10.1371/journal.ppat.1004157 (2014)
Impact Factor: 7.562 - Q1 – D1
215
Monica Chagoyen, José L. Carrascosa, Florencio Pazos, and José M. Valpuesta
Molecular determinants of the ATP hydrolysis asymmetry of the CCT chaperonin complex.
Proteins-Structure Function and Bioinformatics, 82:5, 703–707 doi: 10.1002/prot.24510 (2014)
Impact Factor: 2.627 – Q3
214
Daniel Luque, Josué Gómez-Blanco, Damiá Garriga, Axel F. Brilot, José M. González, Wendy M. Havens, José L. Carrascosa, Benes L. Trus, Nuria Verdaguer, Said A. Ghabrial, and José R. Castón.
Cryo-EM near-atomic structure of a dsRNA fungal virus shows ancient structural motifs preserved in the dsRNA viral lineage.
Proc Natl Acad Sci USA, 111:21, 7641-7646 doi: 10.1073/pnas.1404330111(2014)
Impact Factor: 9.674 - Q1 – D1
213
Aranzazu Cruz-Adalia, Guillermo Ramirez-Santiago, Carmen Calabia-Linares, Mónica Torres-Torresano, Lidia Feo, Marta Galán-Díez, Elena Fernández-Ruiz, Eva Pereiro, Peter Guttmann, Michele Chiappi, Gerd Schneider, José López Carrascosa, Francisco Javier Chichón, Gloria Martínez Del Hoyo, Francisco Sánchez-Madrid, Esteban Veiga.
T cells kill bacteria captured by transinfection from dendritic cells and confer protection in mice.
Cell Host & Microbe, 15:5, 611-622, doi: 10.1016/j.chom.2014.04.006 (2014)
Impact Factor: 12.328 – Q1 – D1
212
Mercedes Hernando-Pérez, Elena Pascual, María Aznar, Alina Ionel, José R. Castón, Antoni Luque, José L. Carrascosa, David Reguera and Pedro J. de Pablo.
The interplay between mechanics and stability of viral cages.
Nanoscale, 6:5, 2702-2709, doi: 10.1039/c3nr05763a (2014)
Impact Factor: 7.394 – Q1 – D1
211
Daniel Luque, Andrés de la Escosura, Joost Snijder, Melanie Brasch, Rebecca J. Burnley, Melissa S. T. Koay, José L. Carrascosa, Gijs J. L. Wuite, Wouter H. Roos, Albert J. R. Heck, Jeroen J. L. M. Cornelissen, Tomás Torres and José R. Castón.
Self-assembly and characterization of small and monodisperse dye nanospheres in a protein cage.
Chemical Science, 5, 575-581, doi: 10.1039/c3sc52276h (2014)
Impact Factor: 9.211 – Q1 – D1
210
Ana Cuervo, Mónica Chagoyen, Mar Pulido-Cid, Ana Camacho and José L Carrascosa.
Structural characterization of T7 tail machinery reveals a conserved tubular structure among other Podoviridae family members and suggests a common mechanism for DNA delivery.
Bacteriophage 3:4, e27011, doi: 10.4161/bact.27011, (2013)
Impact Factor: NA
209
Ana Cuervo, Mar Pulido-Cid, Mónica Chagoyen, Rocío Arranz, Verónica A. González-García, Carmela García-Doval, José R. Castón, José M. Valpuesta, Mark J. van Raaij, Jaime Martín-Benito and José L. Carrascosa.
Structural Characterización of the Bacteriophage T7 Tail Machinery.
Journal of Biological Chemistry, 288: 36, 26290-26299, doi: 10.1074/jbc.M113.491209 (2013)
Impact Factor: 4.600 – Q1
208
María I. Daudén, Jaime Martín-Benito, Juan C. Sanchez-Ferrero, Mar Pulido Cid, José M. Valpuesta and José L. Carrascosa.
Large Terminase Conformational Change Induced by Connector Binding in Bacteriphage T7.
Journal of Biological Chemistry, 288, nº 23, pp., 16998-17007, doi: 10.1074/jbc.M112.448951 (2013)
Impact Factor: 4.600 – Q1
207
Alexander Cartagena, Mercedes Hernando-Pérez, José L. Carrascosa, Pedro J. de Pablo and Arvind Raman.
Mapping in vitro local material properties of intact and disrupted virions at high resolution using multi-harmonic atomic force microscopy.
Nanoscale, 5:1, 4729-4736, doi: 10.1039/c3nr34088k, (2013)
Impact Factor: 6.739 – Q1 –D1
206
Elías Herrero-Galán, María Eugenia Fuentes-Perez, Carolina Carrasco, José M. Valpuesta, José L. Carrascosa, Fernando Moreno-Herrero, J.Ricardo Arias-González.
Mechanical Identities of RNA and DNA Double Helices Unveiled at the Single-Molecule Level.
Journal of American Chemical Society, 135:1, 122-131, doi: 10.1021/ja3054755, (2013)
Impact Factor: 11.444 – Q1 –D1
205
Rocío Arranz, Rocío Coloma, F.Javier Chichón, José Javier Conesa, José L. Carrascosa, José Mª Valpuesta, Juan Ortín y Jaime Martín-Benito.
The Structure of Native Influenza Virion Ribonucleoproteins.
Science, 338:6114, 1634-1637, doi: 10.1126/science.1228172 (2012)
Impact Factor: 31.027 – Q1 –D1
204
Alejandro Peña, Inmaculada Matilla, Jaime Martín-Benito, José Mª Valpuesta, J. L. Carrascosa, Fernando de la Cruz, Elena Cabezón and Ignacio Arechaga.
The Hexameric Structure of a Conjugative VirB4 Protein ATPase Provides New Insights for a Functional and Phylogenetic Relationship with DNA Translocases.
Journal of Biological Chemistry, 287:47, 39925-39932, doi: 10.1074/jbc.M112.413849, (2012)
Impact Factor: 4.651 – Q1
203
José A. Morín, Francisco J. Cao, José M. Valpuesta, José L.Carrascosa, Margarita Salas and Borja Ibarra.
Manipulation of single polymerase-DNA complexes. A mechanical view of DNA unwinding during replication.
Cell Cycle 11:16, 2967-2968, doi: 10.4161/cc.21389, (2012)
Impact Factor: 5.321 – Q1
202
Mercedes Hernando-Pérez, Roberto Miranda, María Aznar and José L.Carrascosa.
Direct Measurement of Phage phi29 Stiffness Provides Evidence of Internal Pressure.
Small, 8:15, 2366-2370, doi: 10.1002/smll.201200664 (2012)
Impact Factor: 7.823 – Q1 –D1
201
Laura Fumagalli, Daniel Esteban-Ferrer, Ana Cuervo, José L.Carrascosa and Gabriel Gomila.
Label-free identification of single dielectric nanoparticles and viruses with ultraweak polarization forces.
Nature Materials, 11:9, 808-816, doi: 10.1038/NMAT3369, (2012)
Impact Factor: 35.749 – Q1 –D1
200
Johann Mertens, María Ibarra, José L.Carrascosa and Javier Tamayo.
Interaction of viral ATPases with nucleotides measured with a microcantilever.
Sensor and Actuators B-Chemical, 171, 263-270, doi:10.1016/j.snb.2012.03.062 (2012)
Impact Factor: 3.535 – Q1
199
Ana Cuervo, José L.Carrascosa.
Bacteriophages: Structure.
eLS John Wiley & Sons, Ltd: Chichester, doi: 10.1002/9780470015902.a0024053 (2012)
198
Lara H. Molero, Iván López-Montero, Ileana Márquez, Sonia Moreno, Marisela Vélez, José L. Carrascosa and Francisco Monroy.
Efficient Orthogonal Integration of the Bacteriophage Ø29 DNA-Portal Connector Protein in Engineered Lipid Bilayers.
ACS Synthetic Biology, 1:9, 414-424, doi: 10.1021/sb3000063, (2012)
197
José A. Morín, Francisco J. Cao, José M. Lázaro, J. Ricardo Arias-González, José M. Valpuesta, José L. Carrascosa, Margarita Salas and Borja Ibarra.
Active DNA unwinding dynamics during processive DNA replication.
Proc Natl Acad Sci USA, 109:21, 8115–8120, doi: 10.1073/pnas.1204759109, (2012)
Impact Factor: 9.737 – Q1 –D1
196
J. Gómez-Blanco, D. Luque, J.M. González, J.L. Carrascosa, C. Alfonso, B.L. Trus, W.M. Havens, S.A. Ghabrial and J.R. Castón
Cryphonectria nitschkei Virus 1 Structure Shows that the Capsid Protein of Chrysoviruses Is a Duplicated Helix-Rich Fold Conserved in Fungal Double-Stranded RNA Viruses.
Journal of Virology, 86:15, 8314-8318, doi: 10.1128/JVI.00802-12, (2012)
Impact Factor: 5.076 – Q1
195
Luque D, González JM, Gómez-Blanco J, Marabini R, Chichón J, Mena I, Angulo I, Carrascosa JL, Verdaguer N, Trus BL, Bárcena J, Castón JR.
Epitope Insertion at the N-Terminal Molecular Switch of the Rabbit Hemorrhagic Disease Virus T=3 Capsid Protein Leads to Larger T=4 Capsids.
Journal of Virology, 86:12, 6470-6480, doi: 10.1128/JVI.07050-11 (2012)
Impact Factor: 5.076 – Q1
194
Ortega-Esteban A, Horcas I, Hernando-Pérez M, Ares P, Pérez-Berná AJ, San Martín C, Carrascosa JL, de Pablo PJ and Gómez-Herrero J.
Minimizing tip-sample forces in jumping mode atomic force microscopy in liquid.
Ultramicroscopy, 114, 56-61, doi: 10.1016/j.ultramic.2012.01.007 (2012).
Impact Factor: 2.470 – Q2
193
Peña A, Gewartowski K, Mroczek S, Cuéllar J, Szykowska A, Prokop A, Czarnocki-Cieciura M, Piwowarski J, Tous C, Aguilera A, Carrascosa JL, Valpuesta JM and Dziembowski A.
Architecture and nucleic acids recognition mechanism of the THO complex, an mRNP assembly factor.
EMBO Journal, 31:6, 1605-1616, doi: 10.1038/emboj.2012.10, (2012)
Impact Factor: 9.822 – Q1 –D1
192
Martinez-Martín D, Carrasco C, Hernando-Perez M, de Pablo PJ, Gomez-Herrero J, Perez R, Mateu MG, Carrascosa JL, Kiracofe D, Melcher J, Raman A.
Resolving structure and mechanical properties at the nanoscale of viruses with frequency modulation atomic force microscopy.
PLoS One, 7:1, e30204, doi: 10.1371/journal.pone.0030204 (2012)
Impact Factor: 3.730 – Q1